Yes, we can!
Rick O. Gilmore
2017-07-26 07:37:54
Acknowledgments

Why this discussion & why now?
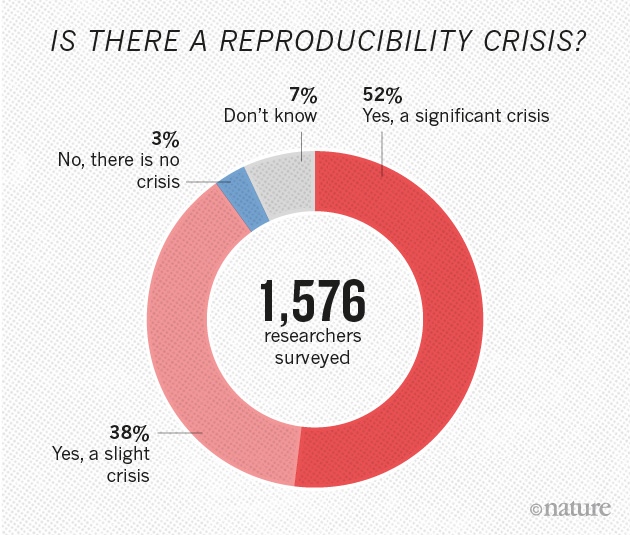
Is there a reproducibility crisis?
- Yes, a significant crisis.
- Yes, a slight crisis.
- No, there is no crisis.
- Don’t know.
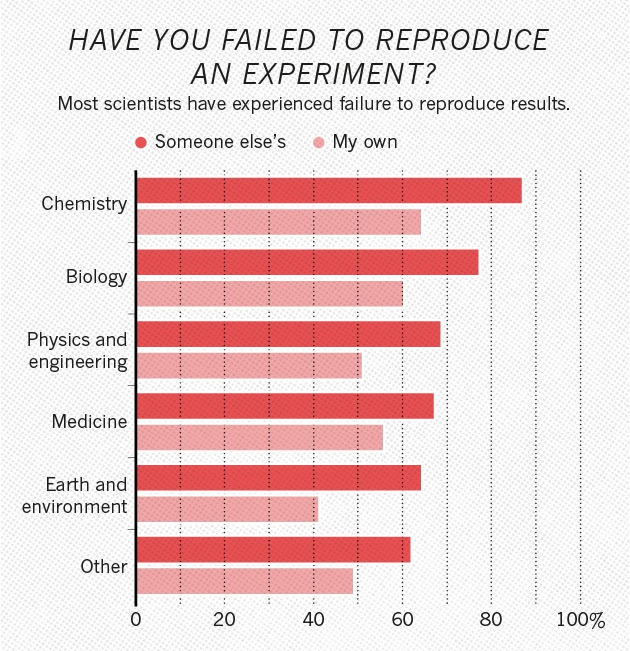
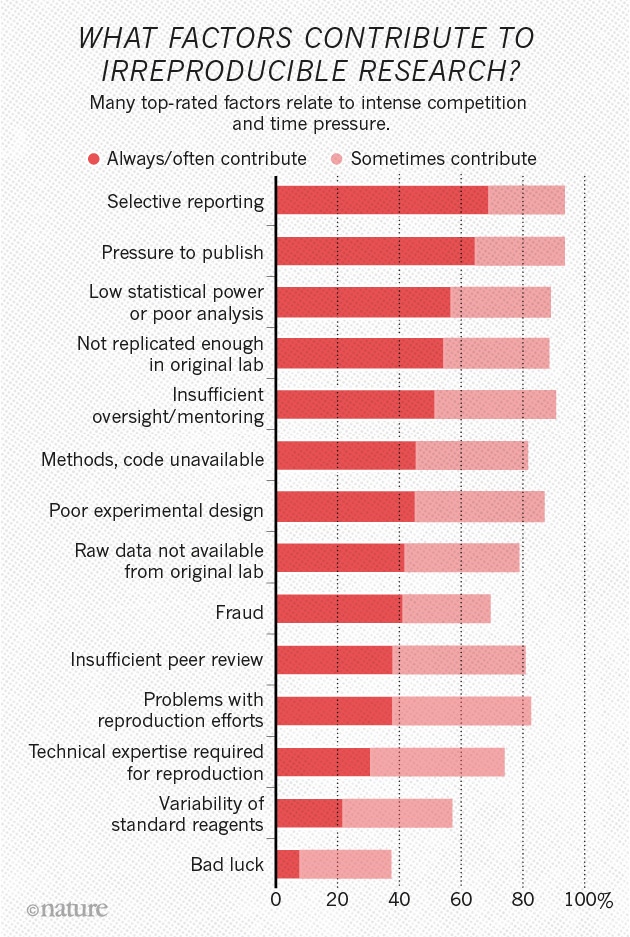
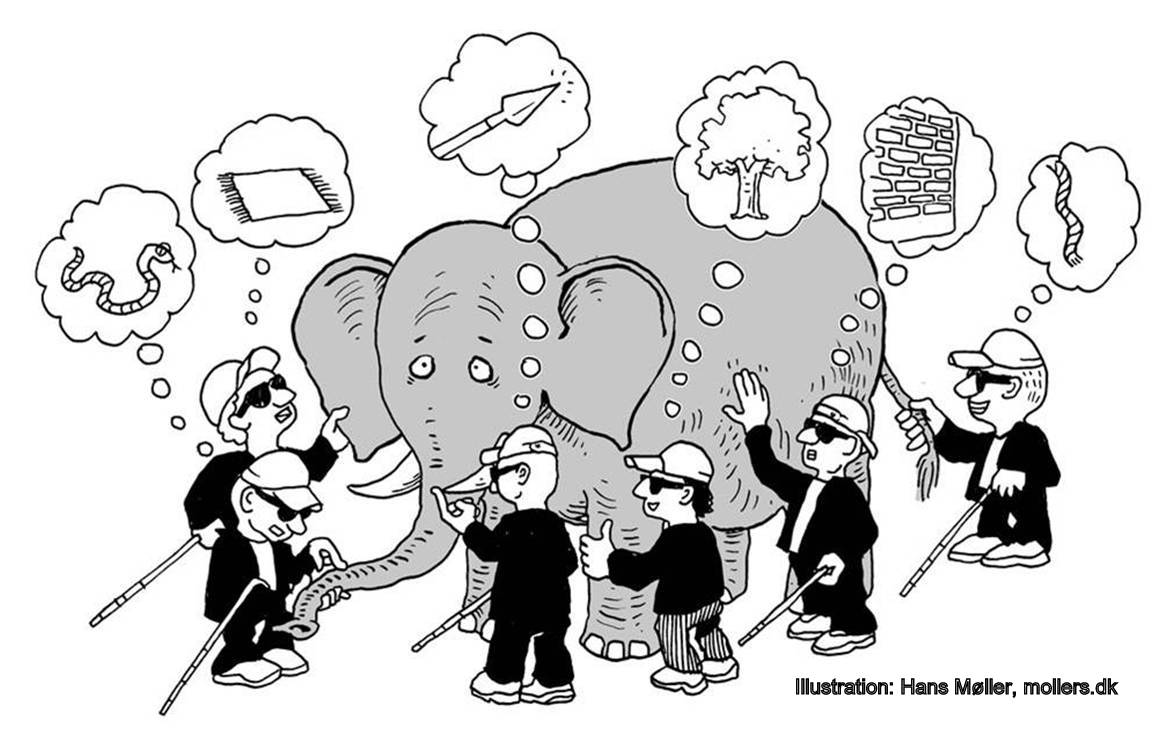
Behavioral science is harder than physics
What’s the more important and lasting contribution?
Our (possibly wrong) findings?
Our (well-curated) data?


Journals +
Repositories =
Structures of knowledge
Journals
- Public face(s)
- Peer review
- Thematic foci
- Consistent formatting, standards
Repositories
Lab, departmental, institutional web sites.Dropbox, Box, Google, etc.- Domain specific, like journals
- Long-term preservation, persistent identifiers
- Foundation of future platforms for discovery
Who owns your data?
you- your institution
- your sponsors (or the taxpayers)
- your participants
- responsible data stewardship
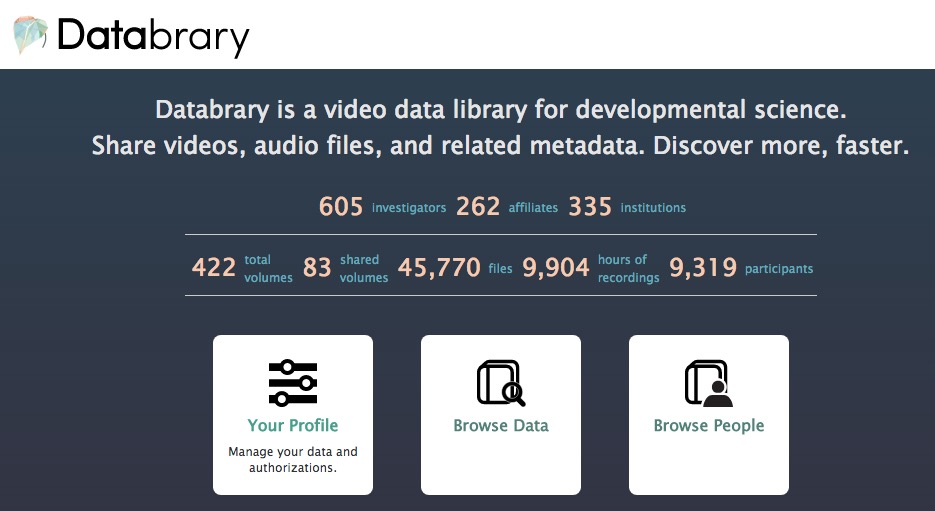
Lessons learned
Identifiable data can be shared
& most participants agree to sharing
Ethical sharing
- Restrict access to researchers
- Institutional (data use & contribution) agreements
- Sharing only with permission
- Permission templates, scripts, videos
- Consistent levels of access
Consistent data management during data collection (active curation) makes sharing easy(er) afterward.
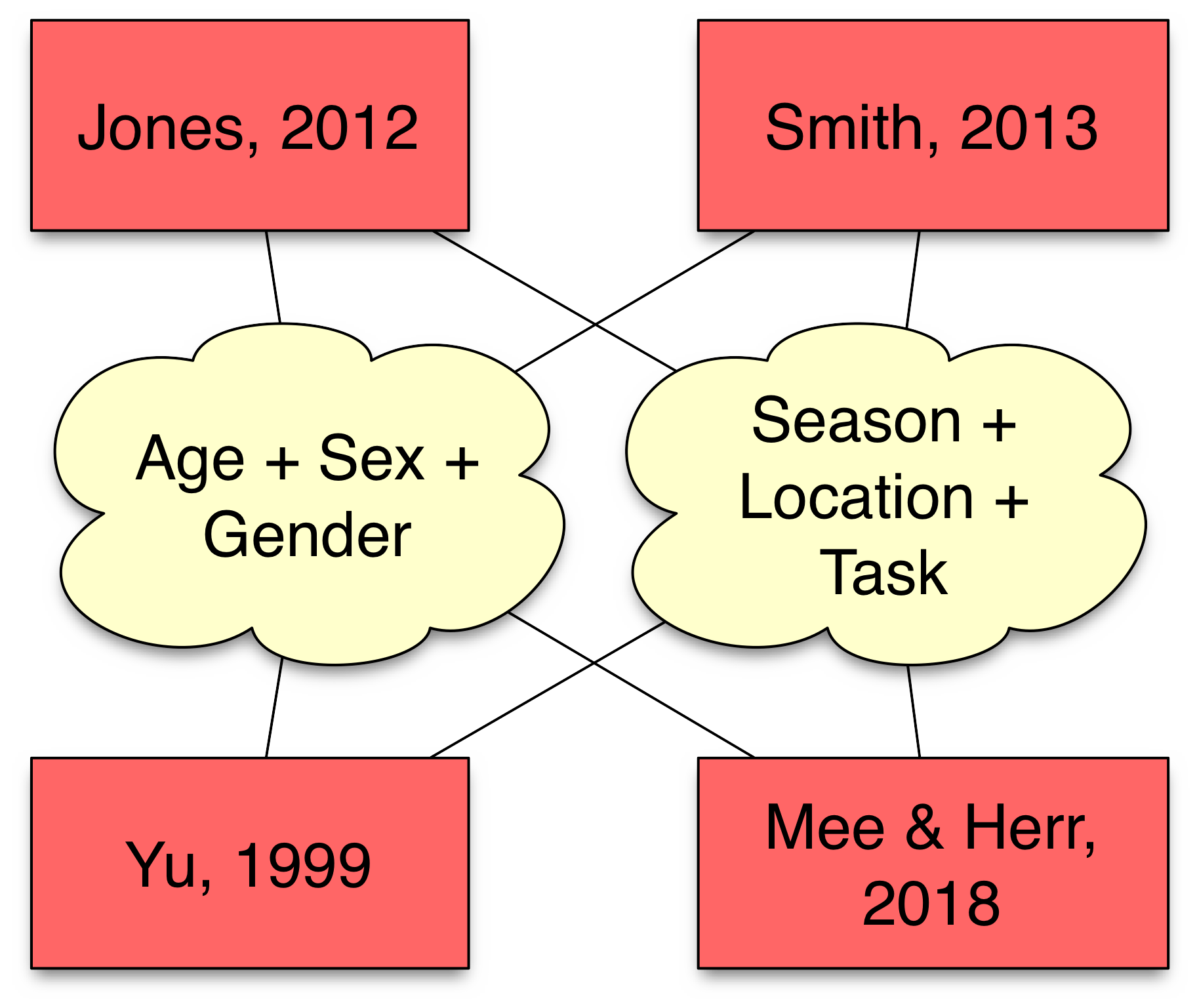
Capturing metadata about people, settings, measures makes data sets searchable, filterable, & more easily reused.
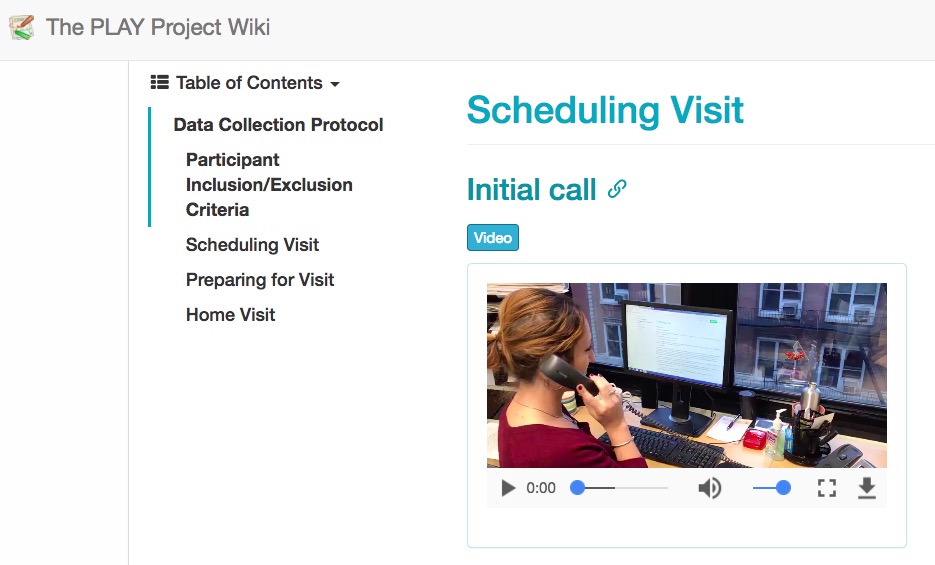
Video as data & documentation
.jpg)


Transparency is good
Accelerating discovery is better
My ‘field of dreams’…
- Link data across studies, measures
- Link across group characteristics, individuals
- Enable searching & filtering by individual characteristics, tasks
- Support web-based data analysis, visualization; open API
- Implement a consistent framework for ethical data sharing
- Enable data aggregation, cloning, provenance tracking
- Support self/active curation
- Link to publications
What’s yours?
Stack
This talk was produced on 2017-07-26 07:37:54 in RStudio 1.0.143 using R Markdown and the reveal.JS framework. The code and materials used to generate the slides may be found at https://github.com/gilmore-lab/aera-workshop-2017-07-26/. Information about the R Session that produced the code is as follows:
## R version 3.4.0 (2017-04-21)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.5
##
## Matrix products: default
## BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## loaded via a namespace (and not attached):
## [1] compiler_3.4.0 backports_1.0.5 magrittr_1.5 rprojroot_1.2
## [5] htmltools_0.3.6 tools_3.4.0 revealjs_0.9 yaml_2.1.14
## [9] Rcpp_0.12.10 stringi_1.1.5 rmarkdown_1.5 knitr_1.16.4
## [13] stringr_1.2.0 digest_0.6.12 evaluate_0.10