Data sharing, research ethics, & scientific reproducibility
Rick Gilmore (rogilmore@psu.edu)
2017-09-28 11:29:37
Preliminaries


About
- Associate Professor of Psychology
- Brain development, perceptual and cognitive development, big data, open science
- Inaugural Director of Human Imaging, Social, Life, & Engineering Sciences Imaging Center (SLEIC)
- Co-founder of the FIRSt Families database
- Co-founder of the Databrary.org data library
Why should we care about data sharing?
Data sharing
- easier than most PIs think
- improves the reproducibility and robustness of research
- maximizes benefits of participation in research
Data sharing…
- can be accomplished for most types of data…
- with advance planning and permission
- increasingly mandatory for funders and journals
- the responsibility of PIs & institutions
Data hoarding
- is (sadly) the norm in some fields (e.g., psychology & neuroscience)
- is problematic without very good reasons
- undermines the reproducibility of research
- wastes resources, time, & energy
Overview
- The reproducibility “crisis” in science
- The “crisis” in psychology
- Challenges to more open data sharing & some solutions
- Shared responsibilities
The reproducibility “crisis”
Is there a reproducibility crisis?
- Yes, a significant crisis
- Yes, a slight crisis
- No crisis
- Don’t know
Methods reproducibility
- Enough details about materials & methods recorded (& reported)
- Same results with same materials & methods

Results reproducibility
- Same results from independent study
Inferential reproducibility
- Same inferences from one or more studies or reanalyses
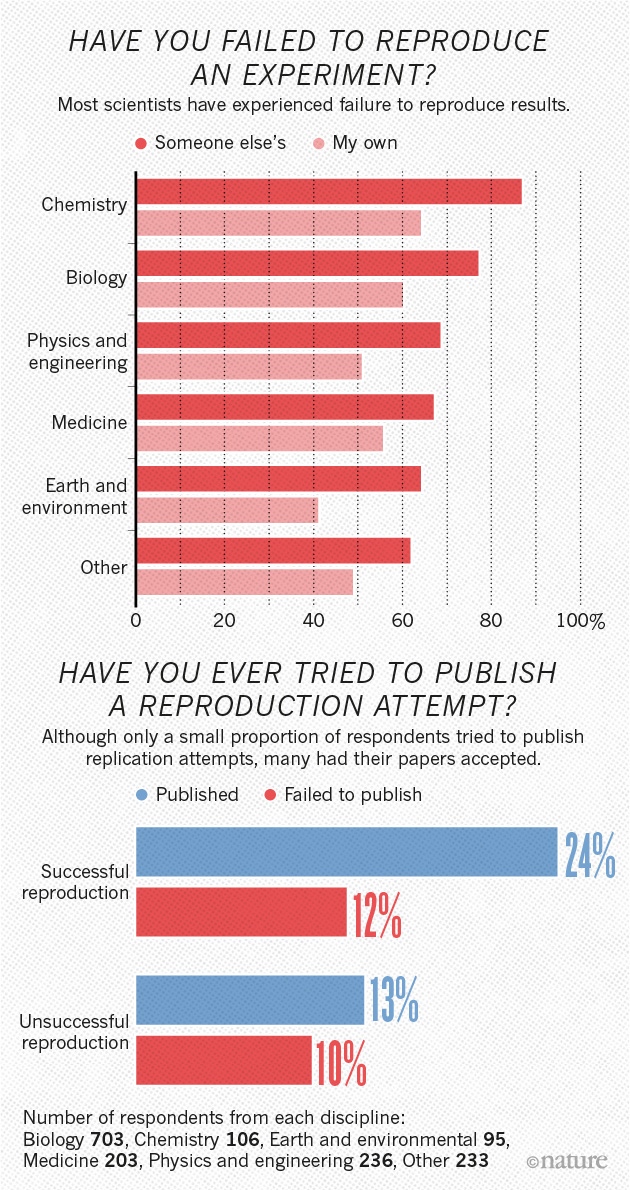
Reproducibility crisis
- Not just psychology
- “Hard” sciences, too
- Data collection to statistical analysis to reporting to publishing
- Ethical urgency

Origins of the crisis in psychology & neuroscience

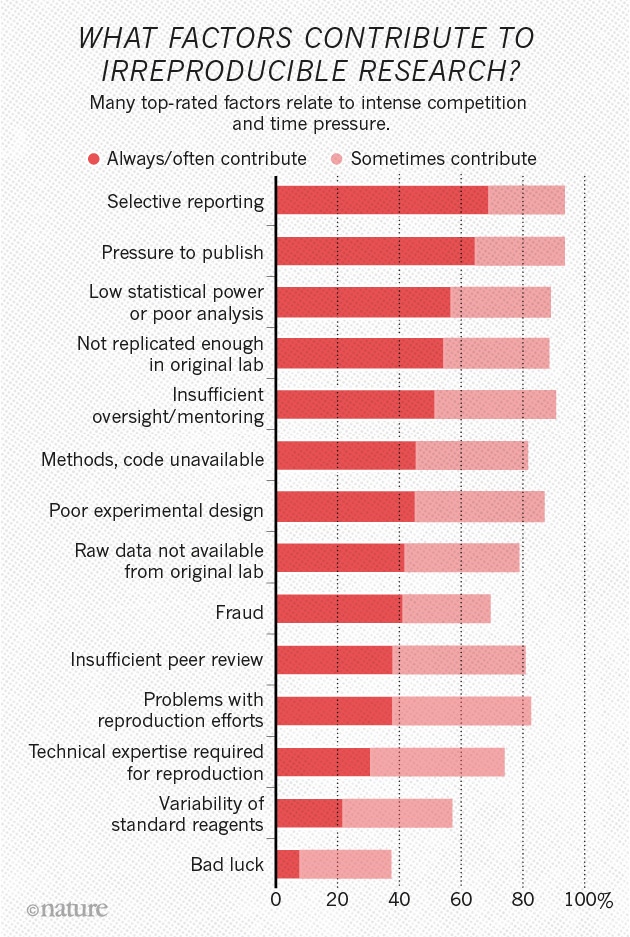
The sin of unreliability
Studies are underpowered
“Assuming a realistic range of prior probabilities for null hypotheses, false report probability is likely to exceed 50% for the whole literature.”
The sin of hoarding…
The sin of corruptibility…
The sin of hurrying…

In our defense…
Psychology is harder than physics
Challenges to sharing
Belmont principles: Beneficence
- Data sharing may pose risk of loss of privacy & confidentiality
- But data sharing increases value, and participation value should be maximal
Belmont principles: Respect for persons
- Data sharing may pose risk of unintended use of data
- So participants should participate in decisionmaking
Belmont principles: Justice
- Costs of research participation should be equitable
- Benefits of research participation should be equitable
How to sharing data ethically
What data are you collecting?
- Personally identifying or sensitive data?
- What risks does data sharing pose?
- How should data be protected?
Where will you share data?
- Researcher/lab/university server
- Supplemental material linked to journal article
- Data repository
Where will you share data?
- Researcher/lab/university server
- Supplemental material linked to journal article
- Data repository
Who will (& should) have access?
- Public
- Community of authorized individuals (researchers)
- Individuals selected by repository
- Individuals selected by data owner
Who will (& should) have access?
- Public
- Community of authorized individuals (researchers)
- Individuals selected by repository
- Individuals selected by data owner
What have participants been told, approved, understood?
- What data collected, what will be shared
- Who will have access
- Where stored, how accessed
- Purposes of use, types of questions
- Shared for how long
Statutory, regulatory, or contractual restrictions?
Benefits (of sharing) vs. risks
- How useful are data?
- How sensitive are data?
- How likely is it that reidentification could be achieved, and by whom?
Assessing disclosure risk
- Reidentification by participants themselves
- Reidentification by insider
- Reidentification by targeted search (nemesis scenario)
- Reidentification by mass matching (dystopian AI scenario)
Mitigating disclosure risk
- Modify data
- Aggregate or censor sensitive variables
- Aggregate or censor secondary identifying variables
- Perturb or add noise to variables
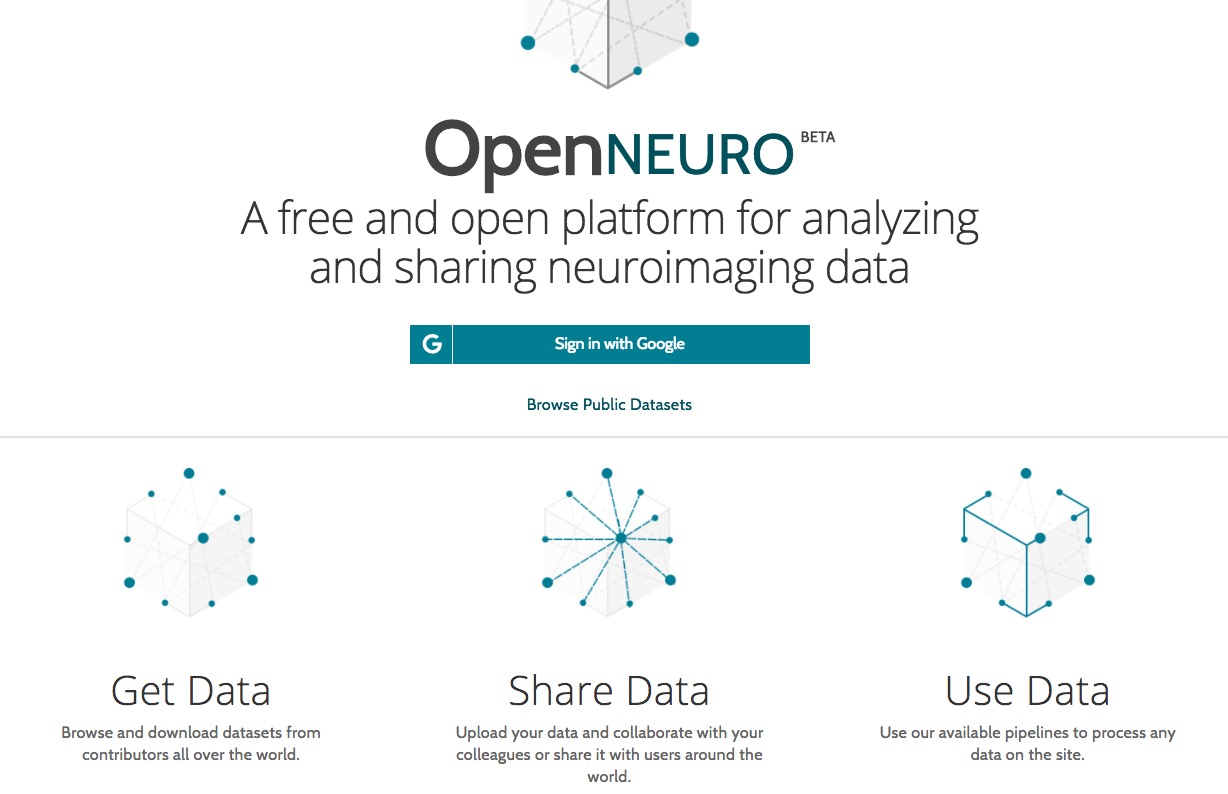
Case study: OpenNeuro.org
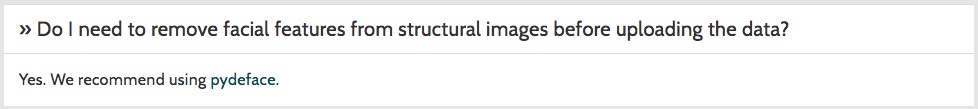
Q: Does modification diminish value for secondary use?
A: “Defacing” MRI: No; “Defacing” video: Yes
Mitigating disclosure risk
- Restrict access
- Data enclaves (e.g., Census data)
- Virtual data enclaves
Mitigating disclosure risk
- Seek explicit sharing permission
- Share unaltered with restricted audiences
- Share identifiable data (video & audio recordings, individual metadata)
- Unaltered to exploit inherent richness for reuse
- Share across studies and researchers
- Not on a study-by-study or PI-by-PI basis
- Share consistently
- Same permission structure
- Same metadata
- Store and serve data securely
- Share identifiable data with permission
- Template sharing permission language
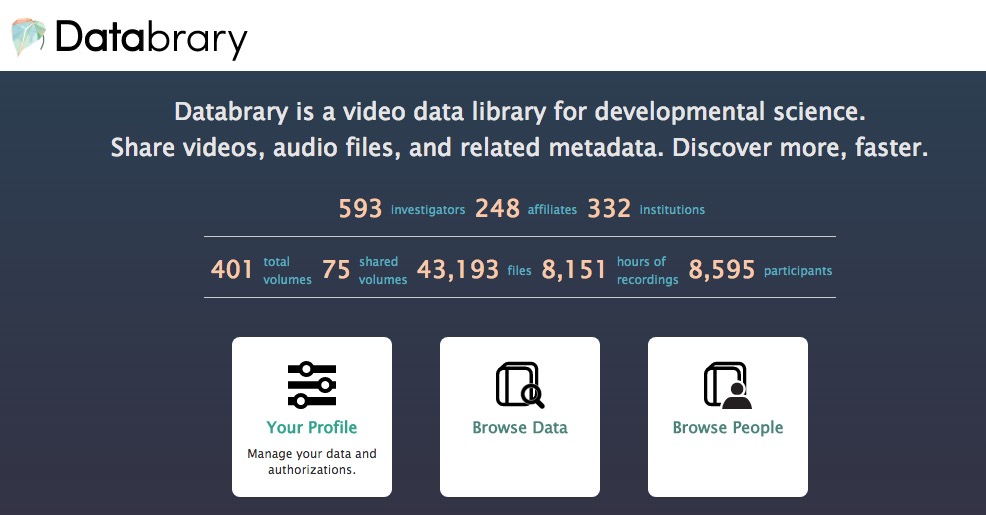
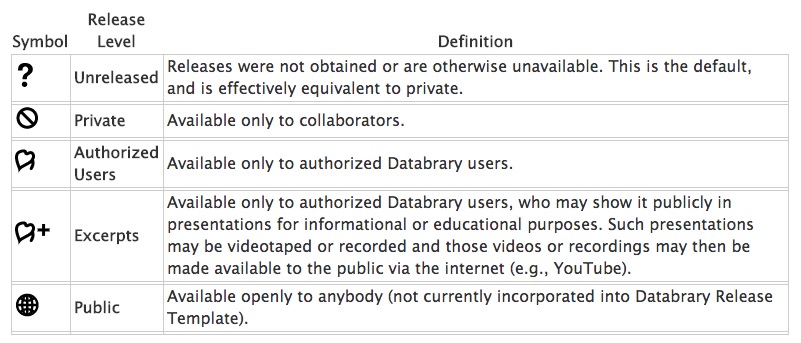
ad hoc video/photo releasesstandard sharing levels
Seeking permission to share
- Open sharing among authorized researchers, not public
- Formal (institution-approved & signed) access agreement
- Laws differ among countries
- Comfort with data sharing varies among institutions
- “Cloud” storage vs. institutionally housed
- Store data (but not share) data for which no permission has been given?
- Role of repositories
- “Check” IRB/ethics training status or delegate responsibility to institutions
My opinion (Gilmore, Kennedy, & Adolph, 2017)
- Biggest obstacle to more open sharing is cultural, not technical
- Planning to share easier than post hoc
- Everyone should be preparing to share
- Share what data, share where, for how long, for what purposes?
- Seeking permission to share should be common practice
- Even if data aren’t sensitive and/or plan to de-identify
- Can’t share (easily) if you don’t ask
Models for data sharing permission
- Separate consent to participate from permission to share
- Same protocol, two different forms
- Two different protocols
- Combine permission to share with consent to participate
Institutional roles
Institutions
- (Often) own data, although PIs don’t always know it
- Have responsibilities under Federal guidelines to help PIs share data
“Investigators are expected to share with other researchers, at no more than incremental cost and within a reasonable time, the primary data, samples, physical collections and other supporting materials created or gathered in the course of work under NSF grants. Grantees are expected to encourage and facilitate such sharing.”
– NSF, Dissemination and Sharing of Research Results, https://www.nsf.gov/bfa/dias/policy/dmp.jsp
Institutions should…
- Partner with PIs
- make planning for data sharing part of grant seeking & reporting process
- ensure data & $ well-managed
Institutions should…
- Make research quality part of criteria for promotion & advancement
- Raise awareness that that reproducibility and research misconduct are genuine concerns
- Strengthen research integrity offices, improve training programs, and develop programs to prevent research misconduct
- Focus especially on mentoring and data management
Compliance office/IRB role
- Data
destruction after X yearssharing forlimitedindefinite period - Share data in
lab websitesecure repository - Encourage seeking permission for
narrowbroad secondary use cases RequireDiscourage reconsent of minors at age of majority unless data highly sensitive- Implement consistent treatment of similar data types
- Encourage use of templates
Progress toward standard templates
- Databrary permission to share templates
- ICPSR language
- Open Brain Consent
- New initiative : Databrary + AERA + ICPSR + QDR + OpenNeuro + …
In summary…
Data sharing
- easier than most PIs think
- improves the reproducibility and robustness of research
- maximizes benefits of participation in research
- increasingly mandatory for funders and journals
Data sharing
- can be accomplished for most types of data with planning & permission
- both PI and institutional responsibility
- upholds scientific ideals
- may be the most ethical thing to do (Brakewood & Poldrack, 2013)

Thank you
rogilmore@psu.edu
gilmore-lab.github.io
gilmore-lab.github.io/psu-sari-2017-09-28/
Stack
This talk was produced on 2017-09-28 in RStudio using R Markdown and the reveal.JS framework. The code and materials used to generate the slides may be found at https://github.com/gilmore-lab/psu-sari-2017-09-28/. Information about the R Session that produced the code is as follows:
## R version 3.4.1 (2017-06-30)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] DiagrammeR_0.9.2 revealjs_0.9
##
## loaded via a namespace (and not attached):
## [1] Rcpp_0.12.12 compiler_3.4.1 RColorBrewer_1.1-2
## [4] influenceR_0.1.0 plyr_1.8.4 bindr_0.1
## [7] viridis_0.4.0 tools_3.4.1 digest_0.6.12
## [10] jsonlite_1.5 viridisLite_0.2.0 gtable_0.2.0
## [13] evaluate_0.10.1 tibble_1.3.3 rgexf_0.15.3
## [16] pkgconfig_2.0.1 rlang_0.1.2 igraph_1.1.2
## [19] rstudioapi_0.6 yaml_2.1.14 bindrcpp_0.2
## [22] gridExtra_2.2.1 downloader_0.4 dplyr_0.7.2
## [25] stringr_1.2.0 knitr_1.17 htmlwidgets_0.9
## [28] hms_0.3 grid_3.4.1 rprojroot_1.2
## [31] glue_1.1.1 R6_2.2.2 Rook_1.1-1
## [34] XML_3.98-1.9 rmarkdown_1.6 ggplot2_2.2.1
## [37] tidyr_0.6.3 purrr_0.2.3 readr_1.1.1
## [40] magrittr_1.5 backports_1.1.0 scales_0.5.0
## [43] htmltools_0.3.6 assertthat_0.2.0 colorspace_1.3-2
## [46] brew_1.0-6 stringi_1.1.5 visNetwork_2.0.1
## [49] lazyeval_0.2.0 munsell_0.4.3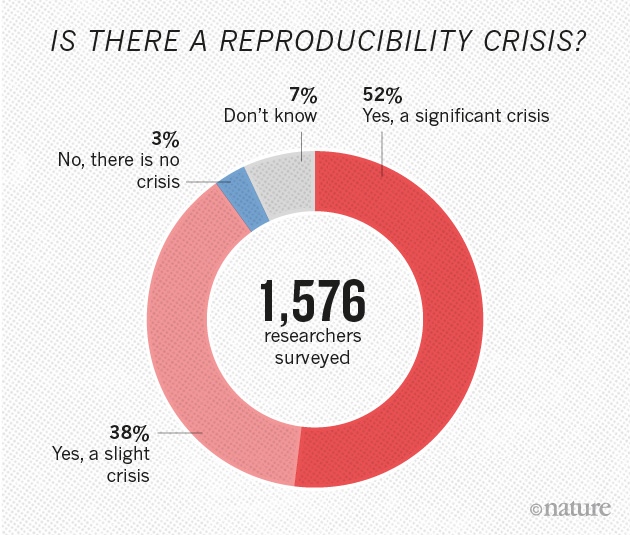 (
( (
( (
(