Reproducibility in Computationally Intensive Behavioral Research
Rick O. Gilmore
2017-09-07 17:03:01
Preliminaries


Overview
- The reproducibility “crisis”
- The “crisis” in psychology
- “Big data” behavioral science is computationally intensive
- Let’s not waste a “good” crisis
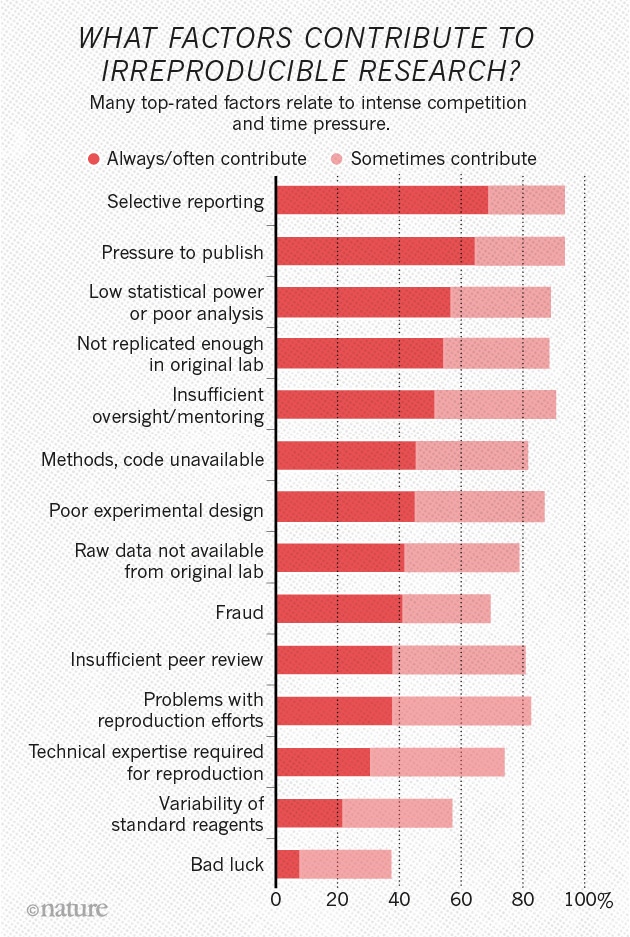
The reproducibility “crisis”
Is there a reproducibility crisis?
- Yes, a significant crisis
- Yes, a slight crisis
- No crisis
- Don’t know
What does “reproducibility” mean?
Methods reproducibility
- Enough details about materials & methods recorded (& reported)
- Same results with same materials & methods

Results reproducibility
- Same results from independent study
Inferential reproducibility
- Same inferences from one or more studies or reanalyses
Reproducibility crisis
- Not just psychology
- “Hard” sciences, too
- Data collection to statistical analysis to reporting to publishing

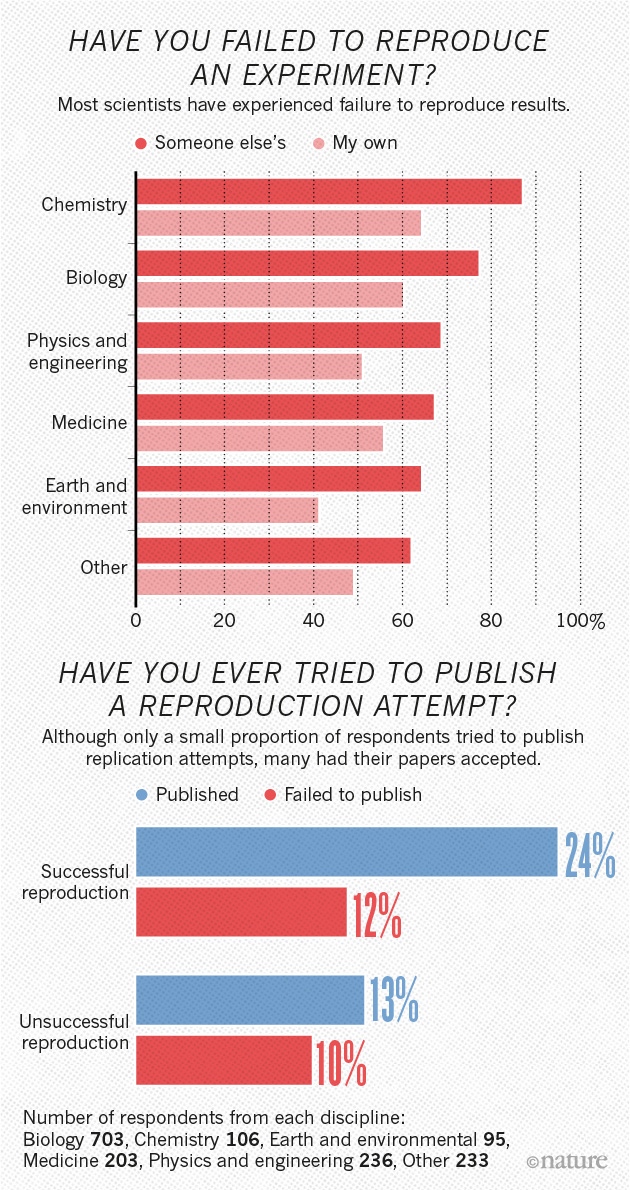
The crisis in psychology

The sin of unreliability
Studies are underpowered
“Assuming a realistic range of prior probabilities for null hypotheses, false report probability is likely to exceed 50% for the whole literature.”
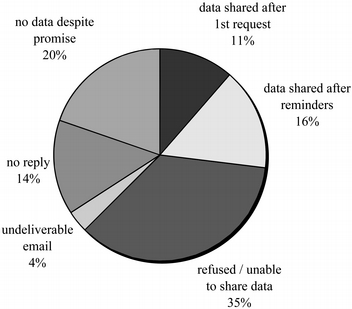
The sin of hoarding…
The sin of corruptibility…
The sin of bias…
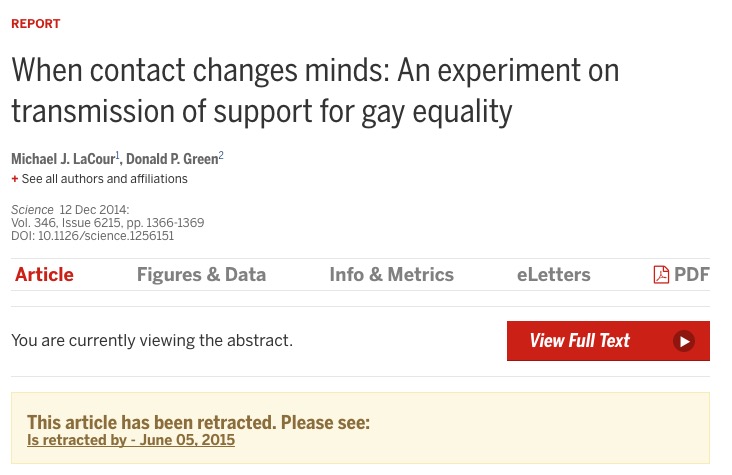
Bem, D.J. (2011). Experimental evidence for anomalous retroactive influences on cognition and affect. Journal of Personality and Social Psychology, 100(3), 407-425.
“This article reports 9 experiments, involving more than 1,000 participants, that test for retroactive influence by”time-reversing" well-established psychological effects so that the individual’s responses are obtained before the putatively causal stimulus events occur."
“We argue that in order to convince a skeptical audience of a controversial claim, one needs to conduct strictly confirmatory studies and analyze the results with statistical tests that are conservative rather than liberal. We conclude that Bem’s p values do not indicate evidence in favor of precognition; instead, they indicate that experimental psychologists need to change the way they conduct their experiments and analyze their data.”
The sin of hurrying…

In our defense…
Behavior multidimensional
Embedded in networks


Humans are diverse
- But much (lab-based) data collected are from Western, Educated Industrialized, Rich, Democratic (WEIRD) populations Henrich et al., 2010
Data sensitive, hard(er) to share
- Protect participant’s identities
- Protect from harm/embarrassment
- Anonymize (effective?) or get permission
Psychology is harder than physics
Big data computation in psychological science
“Mind-reading” in fMRI
A personal example
- How does vision develop?
- Experience
- Input +
- Visually-guided action
- Physical (eye/brain/body) development
Measure (in the lab)
- Behavioral sensitivity
- Brain responses
- At different ages
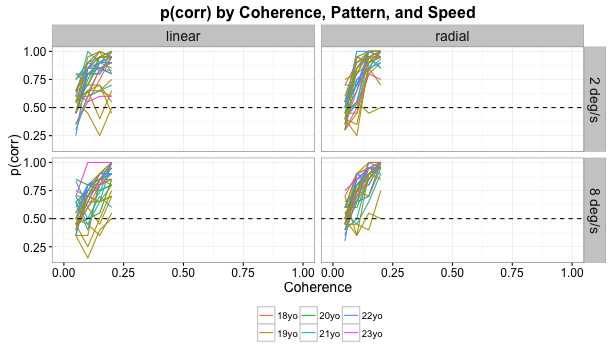
Children’s behavior
Adults’ behavior
Children’s brain responses
Adults’ brain responses
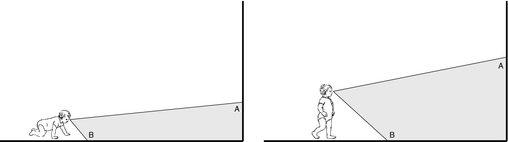
But, what’s the input? The real input?
Frame-by-frame video analysis
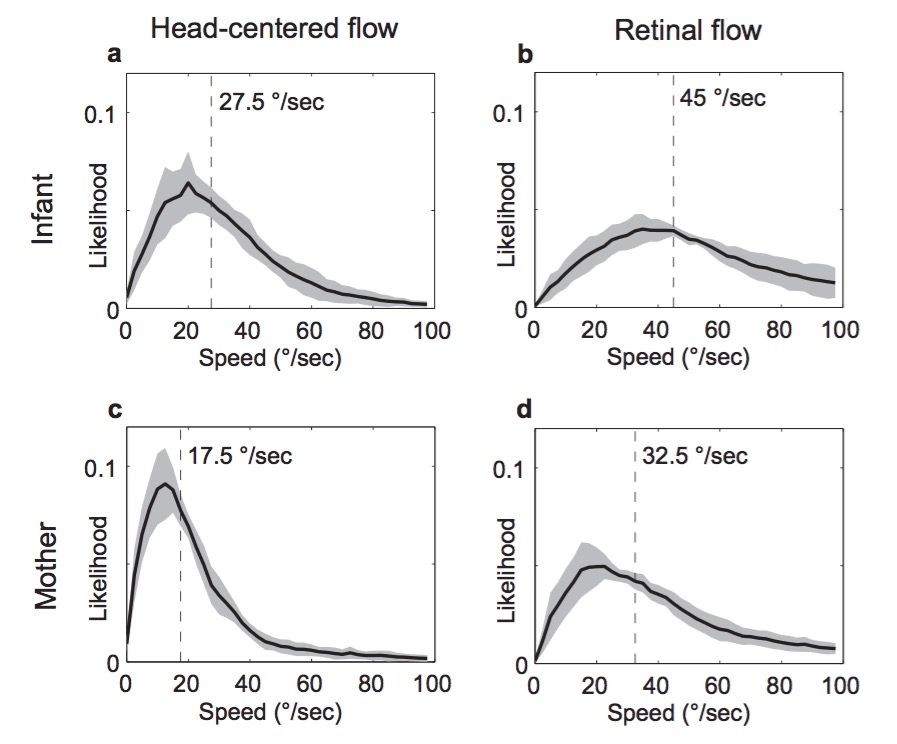
Findings
Findings
- Infant (passengers) experience faster visual speeds than mother
- Controlling for speed of locomotion, environment
- Motion “priors” for infants ≠ mothers
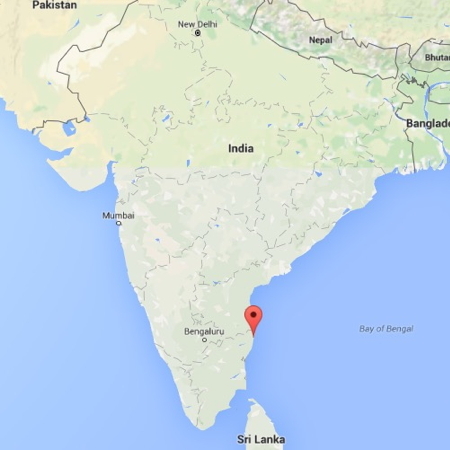
Are “fast” flow speeds common?
| Country | Females | Males | Age (wks) | Coded video Hrs |
|---|---|---|---|---|
| India | 17 | 13 | 3-63 | 3.1 (0.5-6.0) |
| U.S. | 15 | 19 | 4-62 | 4.6 (0.2-7.6) |
Motion speeds - 6 weeks
U.S. | India 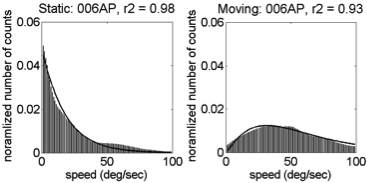
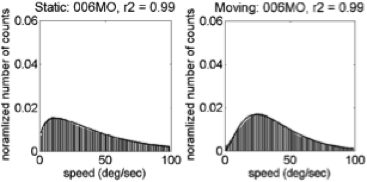 (Gilmore et al, 2015)
(Gilmore et al, 2015)
Motion speeds – 34 weeks
U.S. | India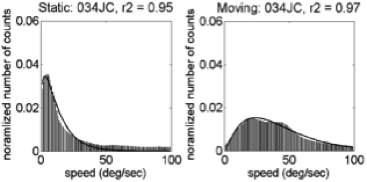
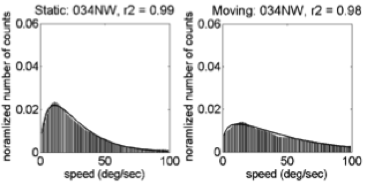 (Gilmore et al, 2015)
(Gilmore et al, 2015)
Motion speeds – 58 weeks
U.S. | India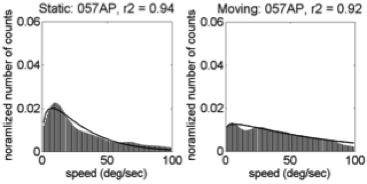
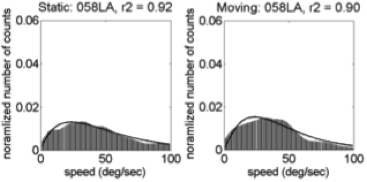 (Gilmore et al, 2015)
(Gilmore et al, 2015)
Linear > radial patterns
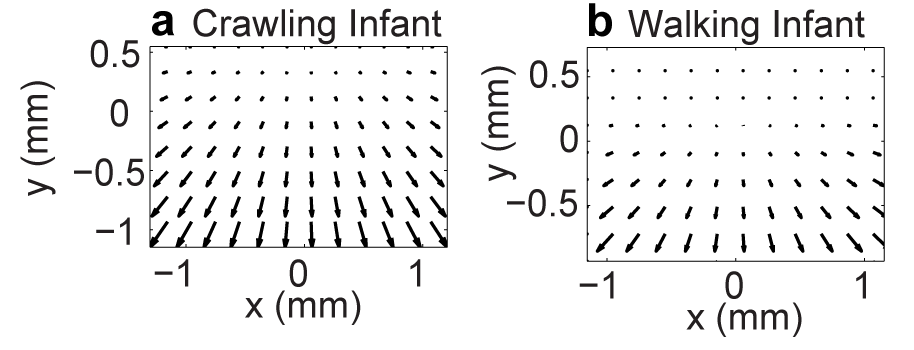
Simulating developmental change
\(\begin{pmatrix}\dot{x} \\ \dot{y}\end{pmatrix}=\frac{1}{z} \begin{pmatrix}-f & 0 & x\\ 0 & -f & y \end{pmatrix} \begin{pmatrix}{v_x{}}\\ {v_y{}} \\{v_z{}}\end{pmatrix}+ \frac{1}{f} \begin{pmatrix} xy & -(f^2+x^2) & fy\\ f^2+y^2 & -xy & -fy \end{pmatrix} \begin{pmatrix} \omega_{x}\\ \omega_{y}\\ \omega_{z} \end{pmatrix}\)
Geometry of environment/observer: \((x, y, z)\) Translational speed: \((v_x, v_y, v_z)\) Rotational speed: \((\omega_{x}, \omega_{y}, \omega{z})\) Retinal flow: \((\dot{x}, \dot{y})\)
Parameters For Simulation
| Parameter | Crawling Infant | Walking Infant |
|---|---|---|
| Eye height | 0.30 m | 0.60 m |
| Locomotor speed | 0.33 m/s | 0.61 m/s |
| Head tilt | 20 deg | 9 deg |
| Geometric Feature | Distance |
|---|---|
| Side wall | +/- 2 m |
| Side wall height | 2.5 m |
| Distance of ground plane | 32 m |
| Field of view width | 60 deg |
| Field of view height | 45 deg |
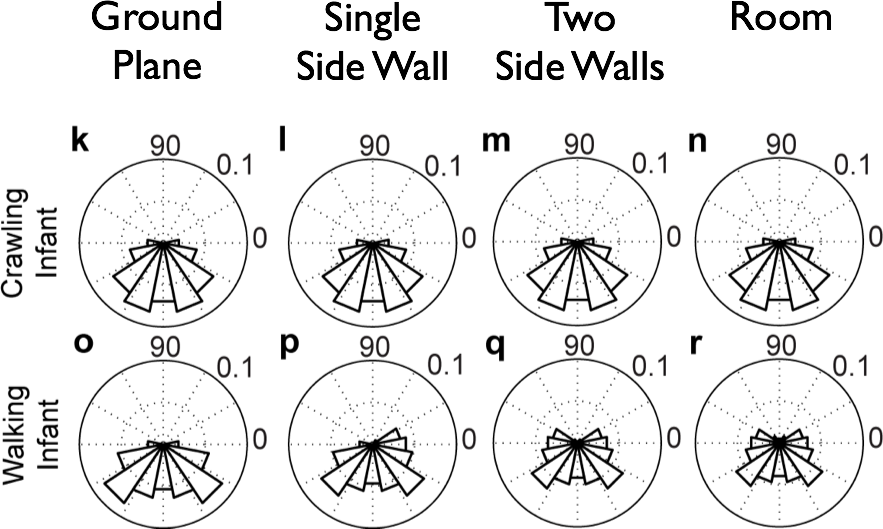
Simulating Flow Fields
Simulated Flow Speeds (m/s)
| Type of Locomotion | Ground Plane | Room | Side Wall | Two Walls |
|---|---|---|---|---|
| Crawling | 14.41 | 14.42 | 14.43 | 14.62 |
| Walking | 9.38 | 8.56 | 7.39 | 9.18 |
Essentials for computationally intensive psychological research
- Computational resources
- Technical expertise
Create reproducible workflows
Kitzes, J., Turek, D., & Deniz, F. (Eds.). (2018). The Practice of Reproducible Research: Case Studies and Lessons from the Data-Intensive Sciences. Oakland, CA: University of California Press. E-book.
Share materials, code, raw data
How Databrary is distinctive
- Open sharing among authorized researchers, not public
- Share identifiable data with permission
- Store, search across, filter among participant & session characteristics
- Active (during study) curation reduces post hoc burden
- Gilmore, Kennedy, & Adolph, 2017
What I’ve learned
Barriers to reproducibility
- Technological
- Cultural

“…psychologists tend to treat other peoples’ theories like toothbrushes; no self-respecting individual wants to use anyone else’s.”
“Reviewers and editors want novel, interesting results. Why would I waste my time doing careful direct replications?”
Any number of researchers I’ve talked with
Tools empower
- RStudio & Project Jupyter and literate programming
- git, GitHub and version control

## Joining multiple datasets
Fancy approach to multiple dataset merge. Joins datasets two at a time from left to right in the list. The result of a two-table join becomes the 'x' dataset for the next join of a new dataset 'y'.```{r data-frame-demo}
df1 <- data.frame(id=1:10, x=rnorm(10), y=runif(10))
df2 <- data.frame(id=1:11, z=rnorm(11), a=runif(11))
df3 <- data.frame(id=2:10, b=rnorm(9), c=runif(9))
Reduce(function(...) { full_join(...) }, list(df1, df2, df3))
```R Markdown

Next generation of scientific publishing
- Lab notebooks that embody literate programming principles
- Close links between data collection, cleaning, analysis, data repositories, preprints, publishers
- Persistent identifiers for research materials, code, & resources
- All published figures, data tables, data sets, analysis code…
Let’s not waste a “good” crisis


Collect & share video as data and documentation

Increase sample sizes

Standardize metadata
- participants (age, gender, race/ethnicity, …)
- settings (times, dates, places)
- measures & tasks

Improve statistical practices
- Automated checking of paper statistics (in American Psychological Association formats) via Statcheck
- Redefine “statistical significance” as \(p<.005\)? (Benjamin et al., 2017)
- Or move away from NHST toward more robust and cumulative practices (Bayesian, CI/effect-size-driven)
Store data, materials, code in repositories
- Data libraries
- Funder, journal mandates for sharing increasing
- But no long-term, stable, funding sources for curation, archiving, sharing
- ArXiv model
- Institutional (Cornell) support
- Subscription
Build platforms for discovery
- Data + analysis
- e.g., PSU’s Biostars


Data from diverse domains



Link measures across people
Web-based data visualization, analysis
Search, filtering by personal characteristics
Curate data & materials as they are generated

Consistent, clear sharing permissions structure
Progress
| Example | Multi-measure | Indiv link/search | Visualize | Self-curate | Permissions |
|---|---|---|---|---|---|
| Databrary | ✔ | ✔ | tabular | ✔ | ✔ |
| Human Proj | ✔ | ✔ | ? | ? | ✔ |
| ICPSR | ✔ | ? | ✔ | ? | ✔ |
| Neurosynth | fMRI BOLD | group data | ✔ | public | NA |
| OpenNeuro | ✔ | ? | ✔ | ✔ | public |
| Open Humans | ✔ | ✔ | ? | ? | ✔ |
| OSF | ✔ | ✔ | public | ||
| WordBank | M-CDI | group metadata | ✔ | ? | public |



Keep in touch
rogilmore@psu.edu
gilmore-lab.github.io
Stack
This talk was produced on 2017-09-07 in RStudio Server Pro using R Markdown and the reveal.JS framework on Penn State’s ACI-ICS RStudio Server Pro instance. The code and materials used to generate the slides may be found at https://github.com/gilmore-lab/aci-ics-2017-09-07/. Information about the R Session that produced the code is as follows:
## R version 3.4.1 (2017-06-30)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] DiagrammeR_0.9.2 revealjs_0.9
##
## loaded via a namespace (and not attached):
## [1] Rcpp_0.12.12 compiler_3.4.1 RColorBrewer_1.1-2
## [4] influenceR_0.1.0 plyr_1.8.4 bindr_0.1
## [7] viridis_0.4.0 tools_3.4.1 digest_0.6.12
## [10] jsonlite_1.5 viridisLite_0.2.0 gtable_0.2.0
## [13] evaluate_0.10.1 tibble_1.3.3 rgexf_0.15.3
## [16] pkgconfig_2.0.1 rlang_0.1.2 igraph_1.1.2
## [19] rstudioapi_0.6 yaml_2.1.14 bindrcpp_0.2
## [22] gridExtra_2.2.1 downloader_0.4 dplyr_0.7.2
## [25] stringr_1.2.0 knitr_1.17 htmlwidgets_0.9
## [28] hms_0.3 grid_3.4.1 rprojroot_1.2
## [31] glue_1.1.1 R6_2.2.2 Rook_1.1-1
## [34] XML_3.98-1.9 rmarkdown_1.6 ggplot2_2.2.1
## [37] tidyr_0.6.3 purrr_0.2.3 readr_1.1.1
## [40] magrittr_1.5 backports_1.1.0 scales_0.5.0
## [43] htmltools_0.3.6 assertthat_0.2.0 colorspace_1.3-2
## [46] brew_1.0-6 stringi_1.1.5 visNetwork_2.0.1
## [49] lazyeval_0.2.0 munsell_0.4.3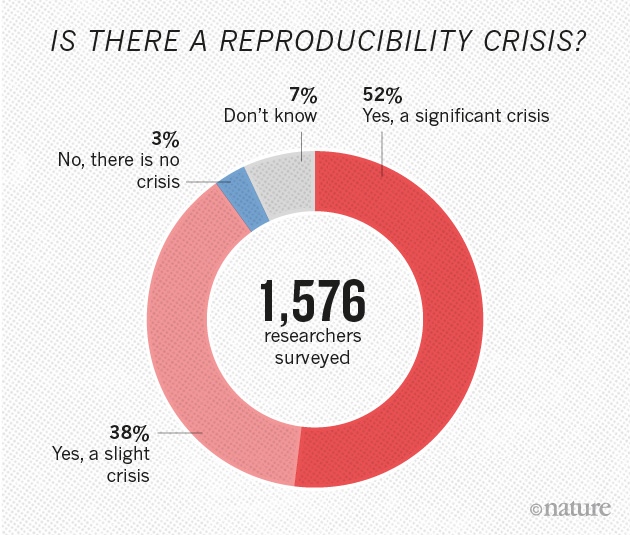 (
( (
( (
(



 (
(
 (
( (
( (
(
 (
( (
( (
( (
( (
(